Intrinsically disordered proteins
Many proteins have been found to be without stable structure in their native states. They are called intrinsically disordered proteins. Their ubiquitous presence undercuts the principle that a protein’s structure determines its function. However, the origins of disorder and its role in protein’s function are still not well understood. This motivated us to look for the general principles that might link protein function and disorder.
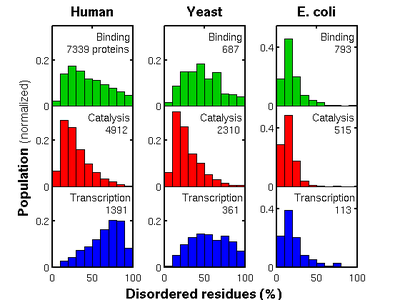
Our comparative genomic analysis showed that proteins' propensity for disorder depends strongly on their function in eukaryotes (figure above): Contrary to the striking bias of catalytic and transcription human proteins to be significantly more ordered and disordered, respectively, disorder is neither strongly favored nor disfavored in binding proteins. These distinctions are still visible in yeast but are less obvious in bacterial genomes such as Escherichia coli, whose proteins are found to be significantly more ordered than those found in eukaryotes across all functional categories.
Based on the above-mentioned preference of disorder among different functional categories, we classified all organisms into three types: (type I) no strong preference for ordered structures in binding proteins but preference for disorder in transcription proteins, among which are human, mouse, zebrafish, chicken, rice, fruit fly, Arabidopsis thaliana, and Dictyostelium discoideum; (type II) no strong preference for ordered structures for either binding or transcription proteins, among which one finds yeast, Schizosaccharomyces pombe, and Caenorhabditis elegans; and (type III) strong preference for ordered structures in both binding and transcription proteins, among which there are E. coli, Bacillus anthracis, and Pseudomonas fluorescens. For catalysis, all genomes show a strong preference for ordered proteins. We note that the smaller bacterial genomes are all type III, whereas eukaryotes are either type I or II, with type I genomes being generally larger than type II.
Our thermodynamic analysis of protein-substrate interaction further showed that evolution may act differentially upon the level of disorder for proteins of different functions: Enzymes are generally weak binders, and they require ordered structures for high catalytic efficiency. Binding Proteins show a broad range of binding affinities, with weak binders requiring ordered structures for high binding efficiency, and with strong binders being able to tolerate disorder. We also showed that disorder can be used to maximize the specificity of promiscuous interactions relevant to transcription and signal transduction.
REFERENCE:
J. Liu, J. R. Faeder, and C. J. Camacho (2009) Toward a quantitative theory of intrinsically disordered proteins and their function. Proc Natl Acad Sci USA 106:19819-19823. (full text)