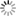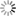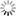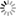Gierasch2009
Notes:
The paper is a review about protein dynamics in signaling transduction. And the main points are:
- Signaling relies on intrinsic dynamic properties of proteins and that proteins relay signals by shifting among different fluctuating energy states in response to one or more inputs.
- Intramolecular signal transmission in PDZ domains seems to arise from a combination of structural changes and dynamic fluctuations. PSD-95 PDZ3 functions as a rigid protein interaction domain.
- The complexity of signal transduction of PDZ domains could be modulated by the linking domains next to PDZ.
- In our study, we don't consider the dynamics of PDZ domain. The dynamics of PDZ won't effect our prediction results. a. peptide binding to PDZ is the signal input and dynamics changing of PDZ is the response to the input. b. The dynamic region may be away from binding site. c. If available, we use bound PDZ structures to do the prediction.
Smock RG, Gierasch LM (2009) Sending signals dynamically. Science 324: 198-203. (pdf)
It is increasingly apparent that signaling relies on the intrinsic dynamic properties of proteins and that proteins relay signals by shifting among different fluctuating energy states in response to one or more inputs.
Just as the ability of a protein to fold is now understood to be best described by an “energy landscape,” which maps the many states that a folding protein can visit as it samples conformational space en route to its native structure, so also is the ability of a signaling protein to respond to signals and pass them on dependent on the features of the energy landscape. The unctional states crucial to signaling are in the lower energy regions of the overall folding landscape (Fig. 1). In both folding and signaling, the conformational states of a protein are populated to varying extents according to their energies, and rates of interconversions between states are governed by the heights of energy barriers between them.
Signals can be transmitted by a shift in the equilibrium population of states for a protein with a rugged energy landscape (7). The “new view” of allostery (11) encapsulates the ideas of dynamics and postulates that the protein populates ensembles of many conformations at all times, fluctuating among these conformations. The interaction with a signaling partner remodels the landscape and consequently shifts the population distribution in such a way as to bias toward a particular downstream event.
PDZ
How does a binding signal alter the energy landscape and lead to a productive signaling response? A central mechanism appears to rely on plasticity within an individual protein signaling domain—in other words, the existence of alternative residue packing networks with coupled dynamic motions. Extensive study of different examples of the widespread PDZ signaling domain has identified potential intramolecular structural and dynamic pathways that appear to connect incoming signals, notably binding to recognition motifs present on upstream partner signaling molecules, to downstream partners. In their many different cellular contexts, PDZ domains function to transduce these binding events into favorable domain-domain assembly of complexes.
The correlation of evolutionary conservation among PDZ domains pointed to a spatially contiguous set of residues as candidates for transmission of functional signals (12), and other methods including NMR dynamics analysis (13, 14), thermal fluctuation analysis of crystal structures (15), and computational simulations of correlated motions (14, 16, 17) found similar networks (Fig. 2). Intriguingly, in one example (the second PDZ domain of human tyrosine phosphatase 1E), the network of residues showing similar binding-induced dynamic changes did not coincide with the set of residues that undergoes structural changes between ligand-bound and free states (14). An in-depth analysis of the dynamics time scales of another PDZ domain (from mouse tyrosine phosphatase BL) showed interconversion between different allosteric states to be relatively slow (microseconds to milliseconds). By contrast, a different PDZ domain (human PSD-95 PDZ3) lacked the dynamic network (18). These results, together with recent analysis using double-mutant cycles, support the notion that different PDZ domains evolve to have different dynamic properties tailored to their specific functions. In this case, PSD-95 PDZ3 functions as a rigid protein interaction domain (19).
Whereas intramolecular signal transmission in PDZ domains seems to arise from a combination of structural changes and dynamic fluctuations, other domains rely more exclusively on dynamics for signaling (20). For example, the phosphotyrosine-binding domain of insulin receptor substrate–1 (IRS-1) binds to the utophosphorylated state of hormone-activated insulin receptor to mediate downstream signaling. A detailed NMR dynamics analysis revealed only subtle conformational changes between free IRS-1 and IRS-1 bound to a phosphotyrosinecontaining peptide (21). Rather, a cluster of dynamically perturbed residues was found to connect the peptide-binding site to a distal surface, where the subsequent downstream signaling interaction was postulated to occur.
Switchable Domain-Domain Interactions Signaling network complexity can be achieved by flexibly linking domains and coupling ignalinduced intradomain structural and dynamic responses to overall domain-domain rearrangements. This strategy accounts for the “Lego”-like evolutionary diversification of signaling pathways by recurrent use of the same switchable modules (40, 41). This hierarchical buildup of signaling complexity is exemplified by the PDZ domains described above, which exist in combinations in many signaling proteins (Fig. 2). Their responses to signals can be altered by their context. For example, the first PDZ domain of mouse phosphotyrosine phosphatase BL (which contains five PDZ domains) modulates the peptide affinity and specificity of its second PDZ domain through an interdomain interaction site that is consistent with the intradomain networks discussed above (42). Similarly, the Drosophila cell polarity protein Par-6 is regulated by the Rho guanosine triphosphatase (GTPase) Cdc42 via binding of Cdc42 to a CRIB domain adjacent to the Par-6 PDZ domain. Cdc42-CRIB activation of the PDZ domain occurs with pronounced rigidification of CRIB and CRIB-PDZ contacts on the opposite side from the PDZ ligand-binding site, consistent with the signaling pathways observed for isolated PDZ domains. This indirect domain-domain rearrangement changes the conformation of the PDZ domain, increasing its affinity for peptide and triggering subsequent cell polarity signaling (43). PDZ conformational switching has also recently been implicated in the INAD scaffold protein of the Drosophila visual photoreceptor (44); this protein was previously thought to be a passive organization template. The new data show that INAD PDZ5 acts as a redox switch, switching an internal cysteine pair to an oxidized state in a lightdependent manner to regulate the binding of target proteins and promote signaling, although the mechanistic details of signal transduction are unclear.